Welcome to RadCamp 2020 - The Marseille Edition
October 5-7, 2020
Location: Online (But spiritually in Marseille, France)
Online content
To facilitate participation by people who may be subject to travel restrictions we will provide a Zoom feed of the instruction, with access to the chat channel for participating in the discussion. Link to come.
Organisers, Instructors, and Facilitators
- Didier Aurelle (Aix-Marseille Université/Mediterranean Institute of Oceanography)
- Alex Baumel (Institut Méditerranéen de Biodiversité et d’Ecologie marine et continent)
- Joana Boavida (CCMAR Center of Marine Sciences (Portugal))
- Isaac Overcast (Institut de Biologie de l’Ecole Normale Superieure)
- Eric Pante (La Rochelle Université)
Binder link
- Get a New>Terminal and execute these commands (takes ~5 minutes):
$ wget https://radcamp.github.io/Marseille2020/binder-reinstall.sh
$ bash binder-reinstall.sh
Schedule
NB: All times are CEST (UTC+2)
| Times | Oct 5 | Oct 6 | Oct 7 |
|---|---|---|---|
| 11:00-12:00 | Introductions/Participant slides | N/A | N/A |
| 14:00-15:20 | Opening Remarks, Setup/Data QC & ipyrad CLI Part 1 | Jupyter notebook intro & ipyrad.analysis api for phylogenetic inference (RAxML) | Assembing and using RAD locus catalogs: exploring the parameter space, Running ipyrad on HPC & other optional analysis cookbooks |
| 15:20-15:30 | Break | Break | Break |
| 15:30-17:00 | ipyrad CLI Part 2 | ipyrad.analysis api for population genetic inference (PCA) | Assisted work session |
Workshop sponsors
This session is funded by the Eccorev Research Federation, the Environmental Genomics Research Group, the IMBE laboratory and the Aix-Marseille University.
Additional ipyrad analysis cookbooks
- Tetrad - A Quartet-based species tree method
- Phylogenetic inference: RAxML
- Demographic analysis: momi2
- Clustering analysis: STRUCTURE
- BPP - Bayesian inference under a multi-species coalescent model
- Bucky - Phylogenetic concordance analysis
- ABBA-BABA - Admixture analysis
RADCamp Marseilles 2020 Group Photo
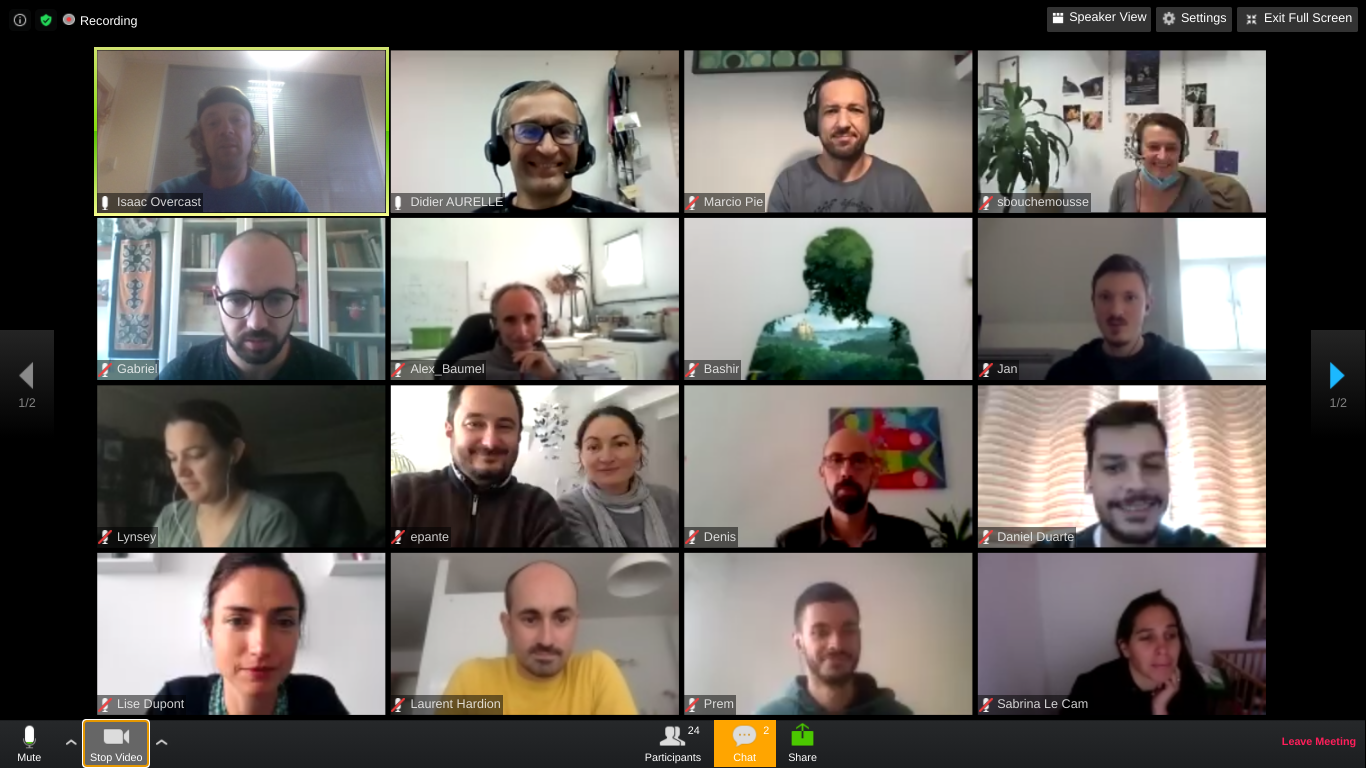
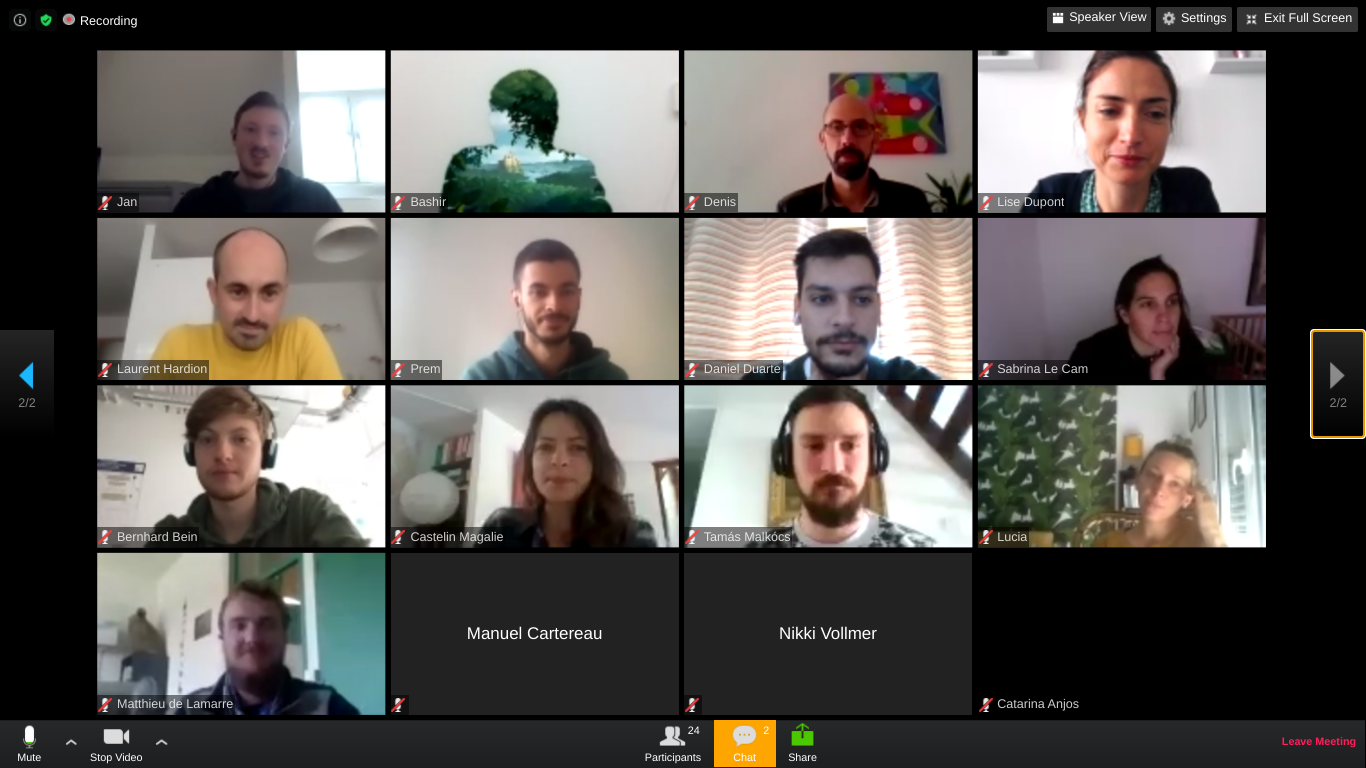
Acknowledgements
RADCamp Marseilles 2020 materials are largely based on materials from previous realizations of the workshop which included important contributions from:
- Deren Eaton
- Mariana Vasconcellos
- Laura Bertola
- Sandra Hoffberg
- Natalia Vasquez
- Francisco Pina Martins